Two distinct sample prep approaches to overcome matrix effect in LC/MS of serum or plasma samples

Introduction
Advancements in electrospray ionization mass spectrometry (ESI-MS) have changed the way identification and quantification of small molecules in biological fluids is conducted. Systems are capable of routinely detecting sub-femtogram levels of analytes. Although these improvements enable low-level detection, ESI-MS is not without limitations, the phenomenon of matrix ionization suppression being a major one. Matrix ionization suppression results from charge competition in the ESI source between components in the sample matrix and the target analytes, and causes diminished, augmented, and irreproducible analyte response.
The Negative Impact of Sample Matrix on LC/MS
Of particular concern with clinical, forensic, and bioanalytical methods is the impact of phospholipids from the matrix. Phospholipids are a major component of cell membranes and notorious for fouling the MS source (Figure 1). They are a major contributor to matrix-induced ionization suppression because they usually co-extract with analytes during sample prep that involves protein precipitation with traditional solid phase extraction (SPE). Additionally, phospholipids often elute in the same timeframe as the analytes from the HPLC column. This co-elution decreases sensitivity which results in increased limits of quantitation and decreased method precision and accuracy. Phospholipids reduce HPLC column lifetime, and their buildup on the column is a problem because they elute erratically, thereby reducing the reproducibility of the method. Excessive gradient elution is needed to wash the HPLC column and the system, which ultimately decreases the sample throughput. The impact of phospholipids on LC/MS experiments has been discussed at length in previous Reporter articles.1-3
Approaches to Reducing Matrix Effect
In this article, two novel techniques to mitigate matrix effects will be discussed. The techniques differ in their approach to discriminate target analytes from the sample matrix. The first technique focuses on the isolation of the sample matrix while allowing the target analytes to remain in the sample solution. The second technique is similar to traditional SPE where target analytes are isolated from the sample matrix. In both cases, the focus will be on matrix reduction from blood plasma or serum samples where phospholipids are the endogenous component of primary concern.
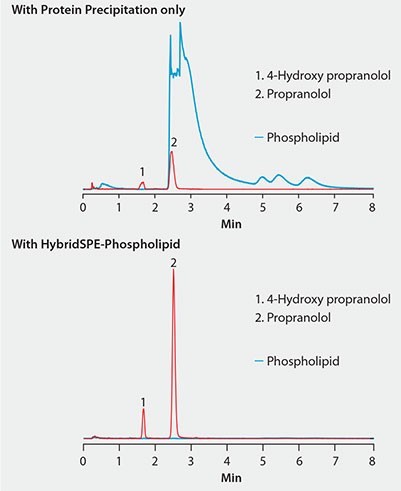
Figure 2. Effectiveness of HybridSPE®-Phospholipid Technique for the Targeted Matrix Isolation Approach to Interference Removal Improved LC/MS response of propranolol and metabolite in plasma after extraction with HybridSPE-PLus compared to protein precipitation
column: Ascentis Express F5, 5 cm × 2.1 mm I.D., 2.7 μm (53567-U)
mobile phase: [A] 2 mM ammonium formate in acetonitrile;
[B] 2 mM ammonium formate in water; (90:10, A:B)
flow rate: 0.4 mL/min
pressure: 1073 psi (74 bar)
column temp.: 35 °C detector: MS, ESI(+) TOF, m/z =100-1000
injection: 2 μL
sample: each compound, 200 ng/mL
system: Agilent 1200SL Rapid Resolution™; 6210 Time of Flight (TOF) MS
Approach 1: Targeted Matrix Isolation
This approach to matrix reduction targets specific components of the sample matrix. In this example, a targeted phospholipid depletion technology (HybridSPE-Phospholipid) is used for the selective isolation of phospholipids from serum or plasma.
HybridSPE-Phospholipid comprises a hybrid zirconia-silica particle packed into 96-well plate or SPE cartridge formats. The electron-deficient empty d-orbitals of the zirconia atoms bond with the electron-rich phosphate groups of the phospholipids via Lewis acid/base interaction. The result is highly efficient phospholipid isolation. Proteins are also removed by precipitation directly in the well plate or tube. Plasma or serum samples are added to the HybridSPE-Phospholipid plate or tube followed by a 3:1 ratio of precipitation solvent. Samples are mixed via draw-dispense or vortex agitation to fully precipitate sample proteins.
Figure 2 shows the same plasma sample processed using standard protein precipitation (top) or phospholipid depletion (bottom) using HybridSPE-Phospholipid. Notice the direct overlap of phospholipids with the target analytes in the protein precipitation sample and concurrent decrease in analyte response compared to the samples processed with the HybridSPE-Phospholipid plate. HybridSPE depleted the phospholipids from the sample, eliminating matrix interference, giving a dramatic increase in analyte response.
Figure 3 shows that the sample processed using protein precipitation resulted in a 75% reduction in response for propranolol due to phospholipid matrix interference. Under these conditions, propranolol eluted in the same region that a large portion of the phospholipids eluted. The error bar for propranolol is much larger due to the irreproducible suppression of the phospholipids. Conversely, samples processed using HybridSPE- Phospholipid demonstrated improved response for propranolol along with much smaller error bars. By selectively isolating the phospholipids, no matrix was introduced onto the analytical column resulting in a
more accurate and precise method.
Approach 2: Targeted Analyte Isolation
The second approach to reduce matrix interference is to target the isolation of analytes while excluding components of the sample matrix. This approach is common in traditional SPE where analyte binding and matrix washing procedures are followed. A continuation of the analyte binding strategy is applied to solid phase micro extraction (SPME) using biocompatible phase chemistries.<sup>4</sup> Figure 4 shows two configurations of biocompatible SPME (bioSPME) fibers currently available from Supelco.
SPME is based upon an equilibrium distribution of the analytes between the solution (serum or plasma, in this case) and the phase layer on the fiber. BioSPME fibers comprise C18-modified silica particles embedded in biocompatible binder. Analytes are desorbed from the fiber using traditional reversed-phase HPLC solvents and analyzed directly by LC/MS. An interesting property of the bioSPME fiber is that the particle binder shields larger biomolecules from adhering to the fiber. This permits the concentration of target analytes on the fiber without coextraction of the sample matrix. The result is a very unique process, whereby sample concentration and sample cleanup are conducted simultaneously. Because bioSPME is not destructive, multiple extractions of the same sample can be performed.
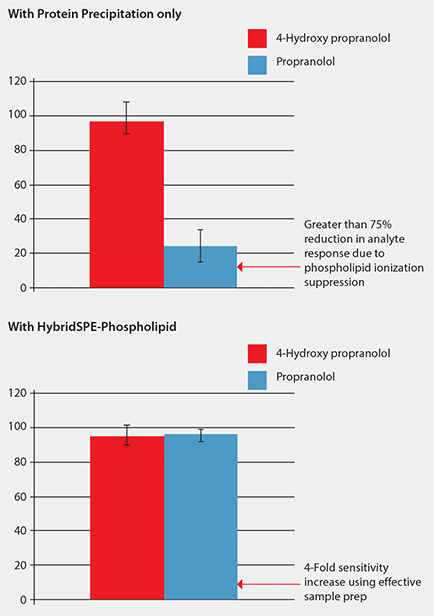
Figure 3. Comparison of Analyte Response (MS) Using Standard Protein Precipitation vs. Phospholipid-Removal Plates (Analysis conditions as in Figure 2. Phospholipid depletion using HybridSPE-PLus plates.)
Figure 5 shows a plasma sample spiked with nine cathinone compounds. In the chromatogram on the top, sample prep consisted of standard protein precipitation. In comparison, the chromatogram on the bottom shows the same sample extracted with the bioSPME fiber. Notice the difference in signal response for both analytes and phospholipids between the two techniques: The bioSPME method gave over twice the analyte response but one-tenth the phospholipid response of the protein precipitation method. This demonstrates the ability of the bioSPME fiber to concentrate analytes from biological samples without interference from matrix components.
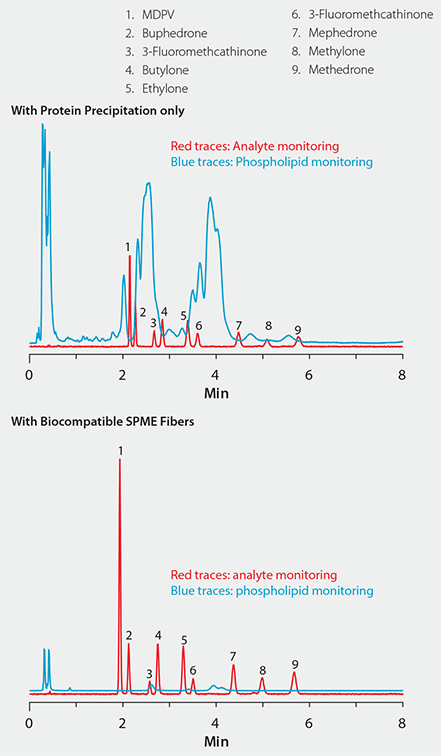
Figure 5. Effectiveness of Biocompatible SPME Technique for the Targeted Analyte Isolation Approach to Interference Removal
column: Ascentis Express HILIC, 10 cm × 2.1 mm I.D., 2.7 μm (53939-U)
mobile phase: 5 mM ammonium formate in 98:2 (v/v) acetonitrile:water
flow rate: 0.6 mL/min pressure: 1842 psi (127 bar)
column temp.: 35 °C detector: MS, ESI(+) TOF, m/z =100-1000 injection: 1 μL
sample: each compound, 50 ng/mL system:
Agilent 1290 Infinity; 6210 Time of Flight (TOF) MS
Summary
Reducing matrix effect is an important consideration when developing an LC/MS method for serum or plasma samples.
This brief report described two distinct sample prep techniques to reduce matrix effect: targeted phospholipid depletion using HybridSPE-Phospholipid and analyte enrichment using biocompatible SPME. Both methods provide effective means to reduce matrix interference that can rob the method of sensitivity, reproducibility, precision, and accuracy.
References
- Aurand, Craig. Understanding, Visualizing and Reducing the Impact of Phospholipid-Induced Ion Suppression in LC/MS; Supelco Reporter Volume 30.2: 10–12.
- Aurand, Craig. Investigating Matrix Interference in Analysis of Antiarrhythmic Cardiac Drugs in Plasma; Supelco Reporter Volume 31.1: 12–15.
- Aurand, Craig. Improvement in LC-MS/MS Analysis of Vitamin D Metabolites in Serum by leveraging Column Selectivity and Effective Sample Prep.; Supelco Reporter Volume 31.1: 16–17.
- Bojko, Barbara; Pawliszyn, Janusz. In vivo and ex vivo SPME: a low invasive sampling and sample preparation tool in clinical bioanalysis. Bioanalysis, 2014, 6(9), 1227–1239.
For more information, or to request a sample, please visit: sigmaaldrich.com/hybridspe (for HybridSPE) and sigmaaldrich.com/biospme (for BioSPME).
Craig R. Aurand