Ligand binding assay technologies: matching the right tool to your bioanalytical challenge

Saloumeh K. Fischer – Genentech, South San Francisco (CA, USA)
Sally obtained her PhD in biochemistry and did a postdoctoral fellowship at Oregon State University (OR, USA) and Lawrence Livermore National Laboratory (LLNL; CA, USA). She was hired as a scientist at LLNL for 2 years, before joining Genentech Research Department as a Scientist in 2000. Sally moved to Development Sciences department in 2003 and is currently an Associate director/Principal Scientist who leads the Immunology and Neurology group in Assay Development and Technology (ADT) with in Bioanalytical Sciences (BAS) department. Her group is responsible for development of assay strategies and qualification/validation of methods to evaluate pharmacokinetics (PK), anti-drug antibodies (ADA) as well as biomarkers in both non-clinical and clinical studies for all Immunology and Neurology indications. This information will be used to inform on dose and dose regimen selection, drug safety, and exposure as well as disease progression, clinical outcome and patient selection.
Keywords: Ligand binding assays (LBAs), Pharmacokinetics (PK), Pharmacodynamic (PD), Antibody therapeutics (AbT), Immunogenicity, biomarkers
Ligand binding assays (LBAs) are critical and widely accepted tools in research and development of protein therapeutics. They are often used for the measurement of pharmacokinetic (PK) and toxicokinetic (TK) biomarkers, such as pharmacodynamic (PD) markers, and immunogenicity or anti-drug antibodies (ADA) during preclinical and clinical development. Enzyme-linked immunosorbent assay (ELISA) has been the gold standard for LBAs, and although ELISAs are still widely used, they pose a number of challenges. ELISAs can be time and reagent intensive, have limited dynamic range and often lack appropriate sensitivity. In addition, matrix tolerance can be especially challenging requiring optimization of the assay for individual matrix [1].
During the past couple of decades, there has been emergence of a number of new platform technologies that have been able to address these challenges. However, no one platform has been able to address all the needed elements. Therefore, it is essential to understand what is being measured and what attributes are needed for a particular measurement so as to determine which platform would be most appropriate for the particular application. Some of the factors that need to be considered include: sensitivity requirements, sample volume availability, multiplexing, dynamic range requirements, platform accessibility, ease of development, outsourcing strategy, platform flexibility and throughput needs. In the case of biomarker assays, it is also important to consider availability of off-the-shelf kits or custom kits by vendors as well as ability to develop home brew assays. The figure below attempts to capture the main feature of some of the newer technologies currently available at a glance.
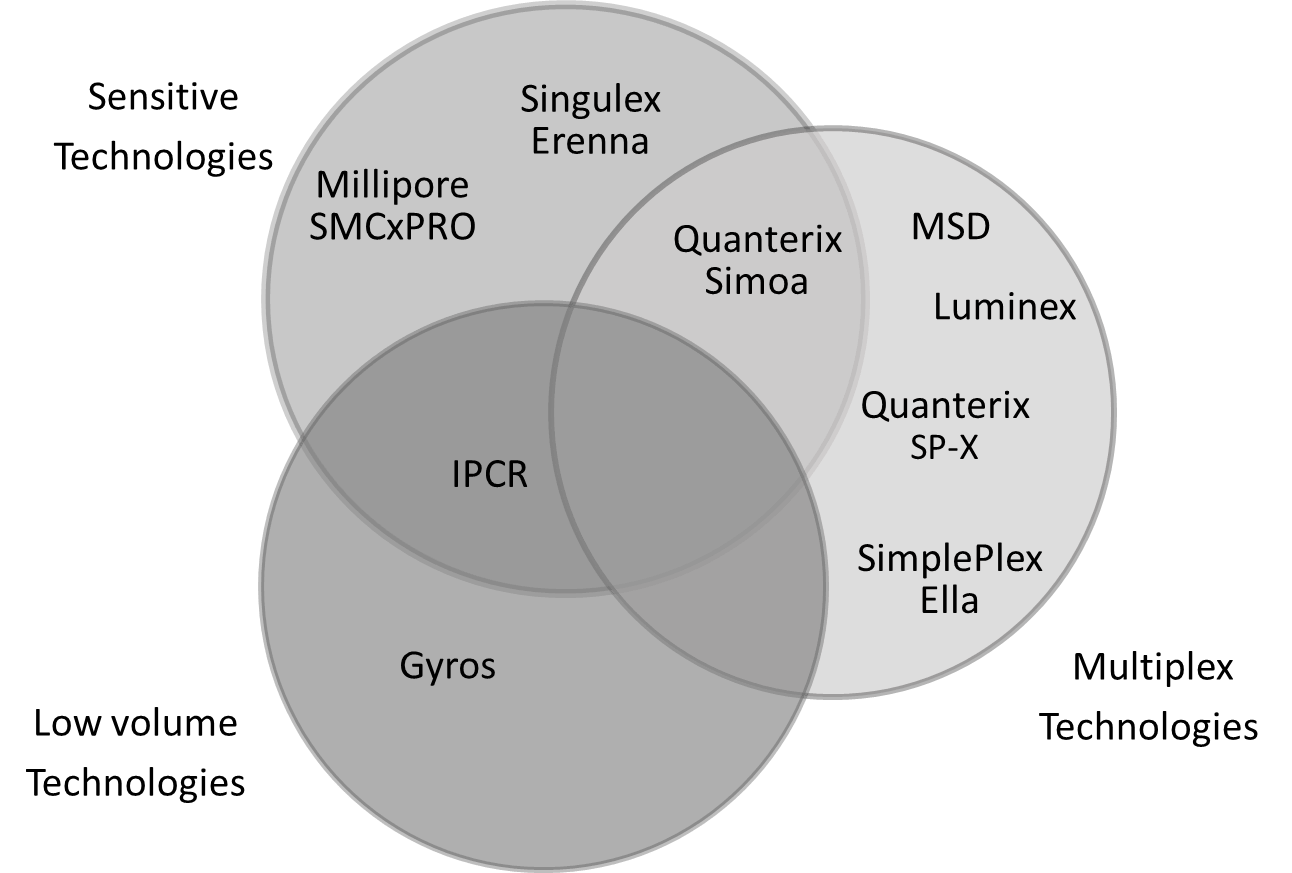 High assay sensitivity is usually required for biomarker assays, as biomarkers are often measured in easily accessible biological fluids such as serum, plasma and CSF, which are used as surrogate for site of disease action. Biomarker levels in these biological matrices can be low at baseline and are often reduced further by treatment necessitating a need for biomarker assays with good sensitivity. The need for sensitivity may also arise for PK measurements. Two examples are, in selection of a safe starting dose and escalation for first in human (FIH) studies where MABLE (minimum anticipated biological effect level) dosing is required or for evaluation of serum PK levels in ocular dosing.
High assay sensitivity is usually required for biomarker assays, as biomarkers are often measured in easily accessible biological fluids such as serum, plasma and CSF, which are used as surrogate for site of disease action. Biomarker levels in these biological matrices can be low at baseline and are often reduced further by treatment necessitating a need for biomarker assays with good sensitivity. The need for sensitivity may also arise for PK measurements. Two examples are, in selection of a safe starting dose and escalation for first in human (FIH) studies where MABLE (minimum anticipated biological effect level) dosing is required or for evaluation of serum PK levels in ocular dosing.
Assay sensitivity in a large part depends on the use of reagents with high affinity, selectivity and specificity for their target. Although incremental improvements in sensitivity may be achieved by optimizing assay conditions, such as buffers, reagent concentrations, and incubation times, significant improvements for ultra-sensitivity (sub-pg/mL–fg/mL) require use of new sensitive detection technologies. Four promising immunoassay platforms in the immunoassay toolbox include Single Molecule Counting (SMC™) from Singulex Inc, SMC (SMCxPro™) from Millipore-Sigma, Single Molecule Arrays (SimoaTM) from Quanterix Corporation, and Immuno-PCR (Imperacer®) from Chimera Biotec GmbH. These platforms have been able to significantly improve immunoassay sensitivity to address the bioanalytical needs for sensitive measurements during biopharmaceutical drug development [2]. Depending on the assay, these platforms are able to achieve pg/mL to fg/mL limits of detection for various protein analytes. Three of these technologies (Simoa, Errena and SMCxPro) achieve their sensitivity by the ability to measure single binding events [3–8]. Immuno-polymerase chain reaction (IPCR) method combines the high specificity of antibody binding with the exponential signal amplification of a downstream quantitative-PCR (qPCR) [9] read out enabling this technology to detect low quantities of bound analytes.
One major challenge in biomarker discovery is disease heterogeneity. New genomic and proteomic studies have revealed that many diseases considered monotypic diseases actually comprise multiple molecular diseases that share a common clinical presentation [10–13]. Therefore, the concept that one biomarker can be predictive in defining a patient population may need to be replaced by an integrative approach that includes multiple biomarkers requiring multiplex technology platforms. For measurement of multiple biomarkers, commercial multiplex kits are often the only option as development of a home brew multiplex assay can be challenging. However, finding quality commercial kits could be challenging. In a recent study, 11 serum biomarkers were assayed using kits from five vendors on three commonly used technologies (Ella, Luminex and Aushon now Quanterix SP-X) [14]. Surprisingly, the results indicated that less than 43% of the assays evaluated were suitable for use. The challenge for the manufacturers of multiplexed kits has been the choice of appropriate combination of biomarkers for multiplexing, since the panel of analytes could include analytes that are present in circulation at very different concentrations. There is also often lack of sensitivity/detectability and specificity for the measurement of low abundant analytes, as well as cross reactivity between reagents in the multiplex kit. These challenges arise because kit manufactures often evaluate biomarker assays in buffer instead of relevant matrix. If they are evaluated in matrix, it is often in a limited number of healthy serum or plasma instead of the disease matrix. The sensitivities reported are often based on buffer standard curves using recombinant material and therefore, an assay could be reported as having very high sensitivity, but in practice, has poor detectability for endogenous analyte in the matrix of interest. Some of these challenges have been addressed by some manufactures and by implementation of new technologies. For example, the Ella platform uses separate nanotubes for each analyte therefore there is no cross-reactivity challenges, as with other multiplex technologies. The Quanterix Simoa and SP-X, as well as MSD [15] have been able to achieve higher detectability with their kits. However, many kits still lack appropriate specificity and suffer from matrix interference, a significant challenge with LBAs.
Matrix interference is defined as ‘the effect of a substance present in the sample that alters the correct value of the result’ [16]. These interferences are due to interactions between constituents in the sample with one or more reagent components and can be analyte-dependent (caused by matrix components) or analyte-independent (caused by assay components). Although interference from heterophilic antibodies has been well documented in literature, it continues to be an underestimated problem that results in erroneous measurements [17–19]. Multiple types of interferences have been identified in serum and plasma capable of significantly impacting measurement in bioanalytical methods. These are often endogenous substances that are natural, polyreactive antibodies or autoantibodies (heterophiles) such as RF, or human anti-animal antibodies (human anti-mouse antibodies (HAMA)). There are several ways to reduce or eliminate these interferences during assay development. One of the strategies is to overcome these interferences using heterophilic antibody inhibitors, such as HeteroBlock, Heterophilic Blocking Reagent (HBR), and Immunoglobulin Inhibiting Reagent (IIR) [20] or elimination of the Fc domain of capture or detection antibodies through use of F(ab′)2 fragments [21] or single-chain fragments (scFv). However, most of these approaches are labor intensive, expensive and not practical for large studies. Other more practical strategies may include sample dilution or reduction of sample incubation time. Sample dilution would compromise assay sensitivity and therefore not a practical option in most instances. Reducing the incubation time or the amount of time the assay reagents are in contact with the matrix has proven effective [1]. One technology that has been very successful in eliminating matrix effects is Gyros. This technology uses a flow through system, which minimizes exposure of reagents to the matrix, and essentially eliminates matrix interference [1]. However, one caveat with this technology is that the capture reagent must have fast on-rate for the assay to work. In addition, Gyros has the advantage of low sample volume, automation and large dynamic range.
LBA technologies have come a long way to address the bioanalytical needs in drug development, although there is still room for improvement. Understanding the advantages and disadvantages of these technologies will allow selection of the most appropriate technology in addressing bioanalytical challenges. Working closely with vendors of these technologies will be key in continued improvement and implementation of these tools.
References:
- Williams K, Erickson R, Fischer SK. Overcoming disease-specific matrix effect in a clinical pharmacokinetic assay using a microfluidic immunoassay technology. Bioanalysis, 9(16), 1207–1216 (2017).
- Fischer SK, Joyce A, Spengler M et al. Emerging technologies to increase ligand binding assay sensitivity. AAPS, J. 17(1), 93–101 (2015).
- Rissin DM, Kan CW, Campbell TG et al. Single-molecule enzyme-linked immunosorbent assay detects serum proteins at subfemtomolar concentrations. Nat. Biotechnol. 28(6), 595–599 (2010).
- Rissin DM, Fournier DR, Piech T et al. Simultaneous detection of single molecules and singulated ensembles of molecules enables immunoassays with broad dynamic range. Anal. Chem. 83(6), 2279–2285 (2011).
- Chang L, Rissin DM, Fournier DR et al. Single molecule enzyme-linked immunosorbent assays: theoretical considerations. J. Immunol. Methods, 378(1–2), 102–115 (2012).
- Shukla R, Santoro J, Bender FC, Laterza OF. Quantitative determination of human interleukin 22 (IL-22) in serum using Singulex-Erenna(R) technology. J. Immunol. Methods, 390(1–2), 30–34 (2013).
- St Ledger K, Agee SJ, Kasaian MT et al. Analytical validation of a highly sensitive microparticle-based immunoassay for the quantitation of IL-13 in human serum using the Erenna immunoassay system. J. Immunol. Methods, 350(1–2), 161–170 (2009).
- Mcfarlin BK, Bowman EM, Gary MA, Stelly S, Tanner EA, Davis A. Using Multiplex and Single-Molecule Bead-Based Technologies to Assess Inflammatory Changes Following Skeletal Muscle Injury. J. Immunol. 200(1 Supplement), 42.48–42.48 (2018).
- Niemeyer CM, Adler M, Wacker R. Detecting antigens by quantitative immuno-PCR. Nat. Protoc. 2(8), 1918–1930 (2007).
- Rossi D, Spina V, Deambrogi C et al. The genetics of Richter syndrome reveals disease heterogeneity and predicts survival after transformation. Blood, 117(12), 3391–3401 (2011).
- Nacht M, Dracheva T, Gao Y et al. Molecular characteristics of non-small cell lung cancer. Proc. Natl. Acad. Sci. U. S. A. 98(26), 15203–15208 (2001).
- Lapointe J, Li C, Higgins JP et al. Gene expression profiling identifies clinically relevant subtypes of prostate cancer. Proc. Natl. Acad. Sci. U. S. A. 101(3), 811–816 (2004).
- Kobel M, Kalloger SE, Boyd N et al. Ovarian carcinoma subtypes are different diseases: implications for biomarker studies. PLoS Med. 5(12), e232 (2008).
- Fischer SK, Carrasco-Triguero M, Hong K et al. Commercial biomarker assays: friend and foe. Bioanalysis, 8(22), 2351–2362 (2016).
- Fu Qin, Zhu Jie, and Van Eyk Jennifer E. Comparison of Multiplex Immunoassay Platforms. Clin. Chem. 56(2): 314–318 (2010).
- Kroll MH, Elin RJ. Interference with clinical laboratory analyses. Clin. Chem. 40(11 Pt 1), 1996–2005 (1994).
- Tate J, Ward G. Interferences in immunoassay. Clin. Biochem. Rev. 25(2), 105–120 (2004).
- Mongolu S, Armston AE, Mozley E, Nasruddin A. Heterophilic antibody interference affecting multiple hormone assays: Is it due to rheumatoid factor? Scand. J. Clin. Lab. Invest. 76(3), 240–242 (2016).
- Bolstad N, Warren DJ, Bjerner J et al. Heterophilic antibody interference in commercial immunoassays; a screening study using paired native and pre-blocked sera. Clin. Chem. Lab. Med. 49(12), 2001–2006 (2011).
- Todd DJ, Knowlton N, Amato M et al. Erroneous augmentation of multiplex assay measurements in patients with rheumatoid arthritis due to heterophilic binding by serum rheumatoid factor. Arthritis, Rheum. 63(4), 894–903 (2011).
- Bjerner J, Nustad K, Norum LF, Olsen KH, Bormer OP. Immunometric assay interference: incidence and prevention. Clin. Chem. 48(4), 613–621 (2002).